Reproduction
Nuclear division is often coordinated with cell division. This generally takes place by mitosis, a process that allows each daughter nucleus to receive one copy of each chromosome. In most eukaryotes, there is also a process of sexual reproduction, typically involving an alternation between haploid generations, wherein only one copy of each chromosome is present, and diploid generations, wherein two are present, occurring through nuclear fusion (syngamy) and meiosis. There is considerable variation in this pattern, however.
Other eukaryotic cells
Eukaryotes are a very diverse group, and their cell structures are equally diverse. Many have cell walls; many do not. Many have chloroplasts, derived from primary, secondary, or even tertiary endosymbiosis; and many do not. Some groups have unique structures, such as the cyanelles of the glaucophytes, the haptonema of the haptophytes, or the ejectisomes of the cryptomonads. Other structures, such as pseudopods, are found in various eukaryote groups in different forms, such as the lobose amoebozoans or the reticulose foraminiferans.
Fungal cell
Fungal cells are most similar to animal cells, with the following exceptions:
- A cell wall that contains chitin
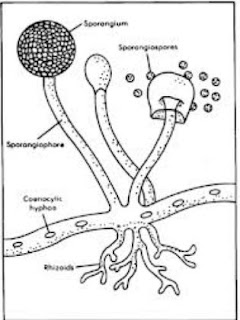
- Less definition between cells; the hyphae of higher fungi have porous partitions called septa, which allow the passage of cytoplasm, organelles, and, sometimes, nuclei. Primitive fungi have few or no septa, so each organism is essentially a giant multinucleate supercell; these fungi are described as coenocytic.
- Only the most primitive fungi, chytrids, have flagella.
Plant cell
Plant cells are quite different from the cells of the other eukaryotic organisms. Their distinctive features are:
- A large central vacuole (enclosed by a membrane, the tonoplast), which maintains the cell's turgor and controls movement of molecules between the cytosol and sap
- A primary cell wall containing cellulose, hemicellulose and pectin, deposited by the protoplast on the outside of the cell membrane; this contrasts with the cell walls of fungi, which contain chitin, and the cell envelopes of prokaryotes, in which peptidoglycans are the main structural molecules
- The plasmodesmata, linking pores in the cell wall that allow each plant cell to communicate with other adjacent cells; this is different from the functionally analogous system of gap junctions between animal cells.
- Plastids, especially chloroplasts that contain chlorophyll, the pigment that gives plants their green color and allows them to perform photosynthesis
- Higher plants, including conifers and flowering plants (Angiospermae) lack the flagellae and centrioles that are present in animal cells
Animal cell
An animal cell is a form of eukaryotic cell that makes up many tissues in animals. The animal cell is distinct from other eukaryotes, most notably plant cells, as they lack cell walls and chloroplasts. They also have smaller vacuoles. Due to the lack of a rigid cell wall, animal cells can adopt a variety of shapes. A phagocytic cell can even engulf other structures.
There are many different cell types. For instance, there are approximately 210 distinct cell types in the adult human body.
Differences among eukaryotic cells
There are many different types of eukaryotic cells, though animals and plants are the most familiar eukaryotes, and thus provide an excellent starting point for understanding eukaryotic structure. Fungi and many protists have some substantial differences, however.
Cell wall
The cells of plants, fungi, and most chromalveolates have a cell wall, a fairly rigid layer outside the cell membrane, providing the cell with structural support, protection, and a filtering mechanism. The cell wall also prevents over-expansion when water enters the cell.
In plants, the major polysaccharides making up the primary cell wall of land plants are cellulose, hemicellulose, and pectin. The cellulose microfibrils are linked via hemicellulosic tethers to form the cellulose-hemicellulose network, which is embedded in the pectin matrix. The most common hemicellulose in the primary cell wall is xyloglucan.
Cytoskeletal structures
Cytoskeletal structures
Many eukaryotes have long slender motile cytoplasmic projections, called flagella, or similar structures called cilia. Flagella and cilia are sometimes referred to as undulipodia, and are variously involved in movement, feeding, and sensation. They are composed mainly of tubulin. These are entirely distinct from prokaryotic flagella. They are supported by a bundle of microtubules arising from a basal body, also called a kinetosome or centriole, characteristically arranged as nine doublets surrounding two singlets. Flagella also may have hairs, or mastigonemes, and scales connecting membranes and internal rods. Their interior is continuous with the cell's cytoplasm.
Microfilamental structures composed by actin and actin binding proteins, e.g., α-actinin, fimbrin, filamin are present in submembraneous cortical layers and bundles, as well. Motor proteins of microtubules, e.g., dynein or kinesin and actin, e.g., myosins provide dynamic character of the network.
Centrioles are often present even in cells and groups that do not have flagella. They generally occur in groups of one or two, called kinetids, that give rise to various microtubular roots. These form a primary component of the cytoskeletal structure, and are often assembled over the course of several cell divisions, with one flagellum retained from the parent and the other derived from it. Centrioles may also be associated in the formation of a spindle during nuclear division.
Significance of cytoskeletal structures is underlined in determination of shape of the cells, as well as their being essential components of migratory responses like chemotaxis and chemokinesis. Some protists have various other microtubule-supported organelles. These include the radiolaria and heliozoa, which produce axopodia used in flotation or to capture prey, and the haptophytes, which have a peculiar flagellum-like organelle called the haptonema.
Mitochondria and plastids
Mitochondria are organelles found in nearly all eukaryotes. They are surrounded by two membranes (each a phospholipid bi-layer), the inner of which is folded into invaginations called cristae, where aerobic respiration takes place. Mitochondria contain their own DNA. They are now generally held to have developed from endosymbiotic prokaryotes, probably proteobacteria. The few protozoa that lack mitochondria have been found to contain mitochondrion-derived organelles, such as hydrogenosomes and mitosomes; and thus probably lost the mitochondria secondarily.
Plants and various groups of algae also have plastids. Again, these have their own DNA and developed from endosymbiotes, in this case cyanobacteria. They usually take the form of chloroplasts, which like cyanobacteria contain chlorophyll and produce organic compounds (such as glucose) through photosynthesis. Others are involved in storing food. Although plastids likely had a single origin, not all plastid-containing groups are closely related. Instead, some eukaryotes have obtained them from others through secondary endosymbiosis or ingestion.
Endosymbiotic origins have also been proposed for the nucleus, for which see below, and for eukaryotic flagella, supposed to have developed from spirochaetes. This is not generally accepted, both from a lack of cytological evidence and difficulty in reconciling this with cellular reproduction.
Internal membrane
Eukaryote cells include a variety of membrane-bound structures, collectively referred to as the endomembrane system. Simple compartments, called vesicles or vacuoles, can form by budding off other membranes. Many cells ingest food and other materials through a process of endocytosis, where the outer membrane invaginates and then pinches off to form a vesicle. It is probable that most other membrane-bound organelles are ultimately derived from such vesicles.
The nucleus is surrounded by a double membrane (commonly referred to as a nuclear envelope), with pores that allow material to move in and out. Various tube- and sheet-like extensions of the nuclear membrane form what is called the endoplasmic reticulum or ER, which is involved in protein transport and maturation. It includes the rough ER where ribosomes are attached to synthesize proteins, which enter the interior space or lumen. Subsequently, they generally enter vesicles, which bud off from the smooth ER. In most eukaryotes, these protein-carrying vesicles are released and further modified in stacks of flattened vesicles, called Golgi bodies or dictyosomes.
Vesicles may be specialized for various purposes. For instance, lysosomes contain enzymes that break down the contents of food vacuoles, and peroxisomes are used to break down peroxide, which is toxic otherwise. Many protozoa have contractile vacuoles, which collect and expel excess water, and extrusomes, which expel material used to deflect predators or capture prey. In higher plants, most of a cell's volume is taken up by a central vacuole, which primarily maintains its osmotic pressure.
Eukaryotes
 |
| Eukaryotes Cell |
Definition
A eukaryote (pron.: /juːˈkæri.oʊt/ or /juːˈkæriət/) is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear envelope, within which the genetic material is carried.[ The presence of a nucleus gives eukaryotes their name, which comes from the Greek ευ (eu, "good") and κάρυον (karyon, "nut" or "kernel"). Most eukaryotic cells also contain other membrane-bound organelles such as mitochondria, chloroplasts and the Golgi apparatus. All large complex organisms are eukaryotes, including animals, plants and fungi. The group also includes many unicellular organisms.
Cell division in eukaryotes is different from that in organisms without a nucleus (Prokaryote). It involves separating the duplicated chromosomes, through movements directed by microtubules. There are two types of division processes. In mitosis, one cell divides to produce two genetically identical cells. In meiosis, which is required in sexual reproduction, one diploid cell (having two instances of each chromosome, one from each parent) undergoes recombination of each pair of parental chromosomes, and then two stages of cell division, resulting in four haploid cells (gametes). Each gamete has just one complement of chromosomes, each a unique mix of the corresponding pair of parental chromosomes.
Eukaryota appears to be monophyletic, and so makes up one of the three domains of life. The two other domains, Bacteria and Archaea, are prokaryotes and have none of the above features. Eukaryotes represent a tiny minority of all living things; even in a human body there are 10 times more microbes than human cells. However, due to their much larger size their collective worldwide biomass is estimated at about equal to that of prokaryotes. They are considered to have first developed approximately 1.6–2.1 billion years ago.
- Cell features
- Differences among eukaryotic cells
Theory that genetic diversity is the adaptive advantage of sex
On the other view, stress is a signal to the cell that it is experiencing a change in the environment to a more adverse condition. Under this new condition, it may be beneficial to produce progeny that differ from the parent in their genetic make up. Among these varied progeny, some may be more adapted to the changed condition than their parents. Meiosis generates genetic variation in the diploid cell, in part by the exchange of genetic information between the pairs of chromosomes after they align (recombination). Thus, on this view, the advantage of meiosis is that it facilitates the generation of genomic diversity among progeny, allowing adaptation to adverse changes in the environment.
However, as also pointed out by Otto and Gerstein, in the presence of a fairly stable environment, individuals surviving to reproductive age have genomes that function well in their current environment. They raise the question of why such individuals should risk shuffling their genes with those of another individual, as occurs during meiotic recombination? Considerations such as this have led many investigators to question whether genetic diversity is the adaptive advantage of sex.
Theory that DNA repair is the adaptive advantage of meiosis
Stress is, however, a general concept.
What is it specifically about stress that needs to be overcome by meiosis? And
what is the specific benefit provided by meiosis that enhances survival under stressful conditions?
Again there are two contrasting theories. In one theory, meiosis is primarily an adaptation for repairing DNA damage. Environmental stresses often lead to oxidative stress within the cell, which is well known to cause DNA damage through the production of reactive forms of oxygen, known as reactive oxygen species (ROS). DNA damages, if not repaired, can kill a cell by blocking DNA replication, or transcription of essential genes.
When only one strand of the DNA is damaged, the lost information (nucleotide sequence) can ordinarily be recovered by repair processes that remove the damaged sequence and fill the resulting gap by copying from the opposite intact strand of the double helix. However, ROS also cause a type of damage that is difficult to repair, referred to as double-strand damage. One common example of double-strand damage is the double-strand break. In this case, genetic information (nucleotide sequence) is lost from both strands in the damaged region, and proper information can only be obtained from another intact chromosome homologous to the damage chromosome. The process that the cell uses to accurately accomplish this type of repair is called recombinational repair.
Meiosis is distinct from mitosis in that a central feature of meiosis is the alignment of homologous chromosomes followed by recombination between them. The two chromosomes which pair are referred to as non-sister chromosomes, since they did not arise simply from the replication of a parental chromosome. Recombination between non-sister chromosomes at meiosis is known to be a recombinational repair process that can repair double-strand breaks and other types of double-strand damage. In contrast, recombination between sister chromosomes cannot repair double-strand damages arising prior to the replication which produced them. Thus on this view, the adaptive advantage of meiosis is that it facilitates recombinational repair of DNA damage that is otherwise difficult to repair, and that occurs as a result of stress, particularly oxidative stress. If left unrepaired, this damage would likely be lethal to gametes and inhibit production of viable progeny.
Even in multicellular eukaryotes, such as humans, oxidative stress is a problem for cell survival. In this case, oxidative stress is a byproduct of oxidative cellular respiration occurring during metabolism in all cells. In humans, on average, about 50 DNA double-strand breaks occur per cell in each cell generation. Meiosis, which facilitates recombinational repair between non-sister chromosomes, can efficiently repair these prevalent damages in the DNA passed on to germ cells, and consequently prevent loss of fertility in humans. Thus on the theory that meiosis arose from bacterial transformation, recombinational repair is the selective advantage of meiosis in both single celled eukaryotes and muticellular eukaryotes, such as humans.
Stress induces sex in bacteria
Bacterial sex (transformation) also appears to be an adaptation to stress. For instance, transformation occurs near the end of logarithmic growth, when amino acids become limiting in Bacillus subtilis, or in Haemophilus influenzae when cells are grown to the end of logarithmic phase. In Streptococcus mutans and other streptococci, transformation is associated with high cell density and biofilm formation. In Streptococcus pneumoniae, transformation is induced by the DNA damaging agent mitomycin C. These, and other, examples indicate that bacterial transformation, like eukaryote meiosis in protists, is an adaptation to stressful conditions. This observation suggests that the natural selection pressures maintaining meiosis in protists are similar to the selective pressures maintaining bacterial transformation. This similarity further indicates continuity, rather than a gap, in the evolution of sex from bacteria to eukaryotes.
Stress induces the eukaryotic sexual cycle in protists
Abundant evidence indicates that facultative sexual protists tend to undergo sexual reproduction under stressful conditions. For instance, the budding yeast Saccharomyces cerevisiae reproduces mitotically (asexually) as diploid cells when nutrients are abundant, but switches to meiosis (sexual reproduction) under starvation conditions. The unicellular green alga, Chlamydomonas reinhardi grows as vegetative cells in nutrient rich growth medium, but depletion of a source of nitrogen in the medium leads to gamete fusion, zygote formation and meiosis. The fissioning yeast Schizosaccharomyces pombe, treated with H2O2 to cause oxidative stress, substantially increases the proportion of cells which undergo meiosis. The simple multicellular eukaryote Volvox carteri undergoes sex in response to oxidative stress or stress from heat shock. These examples, and others, indicate that, in protists and simple multicellular eukaryotes, meiosis is an adaptation to deal with stress.
Sharing of components during the evolution of meiosis and mitosis
On the view that meiosis arose from bacterial transformation, during the early evolution of eukaryotes, mitosis and meiosis could have evolved in parallel, with both processes using common molecular components. On this view, mitosis evolved from the molecular machinery used by bacteria for DNA replication and segregation, and meiosis evolved from the bacterial sexual process of transformation, but meiosis also made use of the evolving molecular machinery for DNA replication and segregation.
Theory that meiosis evolved from mitosis
Mitosis is the process in eukaryotes for duplicating chromosomes and segregating each of the two copies into each of the two daughter cells upon somatic cell division (that is, during all cell divisions in eukaryotes, except those involving meiosis that give rise to haploid gametes). In mitosis, chromosome number is ordinarily not reduced. The alternate theory on the origin of meiosis is that meiosis evolved from mitosis. On this theory, early eukaryotes evolved mitosis first, but lacked meiosis and thus had not yet evolved the eukaryotic sexual cycle. Only after mitosis became established did meiosis and the eukaryotic sexual cycle evolve. The fundamental features of meiosis, on this theory, were derived from mitosis.
Support for the idea that meiosis arose from mitosis is the observation that some features of meiosis, such as the meiotic spindles that draw chromosome sets into separate daughter cells upon cell division, and processes regulating cell division employ the same, or similar, molecular machinery as employed in mitosis.
However, there is no compelling evidence for a period in the early evolution of eukaryotes during which meiosis and accompanying sexual capability was suspended. Presumably such a suspension would have occurred while the evolution of mitosis proceeded from the more primitive chromosome replication/segregation processes in ancestral bacteria until mitosis was established.
In addition, as noted by Wilkins and Holliday, there are four novel steps needed in meiosis that are not present in mitosis. These are: (1) pairing of homologous chromosomes,
(2) extensive recombination between homologs;
(3) suppression of sister chromatid separation in the first meiotic division; and
(4) avoiding chromosome replication during the second meiotic division.
They note that the simultaneous appearance of these steps appears to be impossible, and the selective advantage for separate mutations to cause these steps is problematic, because the entire sequence is required for reliable production of a set of haploid chromosomes.
Theory that meiosis evolved from bacterial sex(transformation)
In prokaryotic sex, DNA from one bacterium is released into the surrounding medium, is then taken up by another bacterium and its information integrated into the DNA of the recipient bacterium. This process is called transformation. One theory on how meiosis arose is that it evolved from transformation. By this view, the evolutionary transition from prokaryotic sex to eukaryotic sex was continuous.
Transformation, like meiosis, is a complex process requiring the function of numerous gene products. The ability to undergo natural transformation among bacterial species is widespread. At least 67 prokaryote species (in seven different phyla) are known to be competent for transformation. A key similarity between bacterial sex and eukaryotic sex is that DNA originating from two different individuals (parents) join up so that homologous sequences are aligned with each other, and this is followed by exchange of genetic information (a process called genetic recombination). After the new recombinant chromosome is formed it is passed on to progeny.
When genetic recombination occurs between DNA molecules originating from different parents, the recombination process is catalyzed in prokaryotes and eukaryotes by enzymes that have similar functions and that are evolutionarily related. One of the most important enzymes catalyzing this process in bacteria is referred to as RecA, and this enzyme has two functionally similar counterparts that act in eukaryotic meiosis, Rad51 and Dmc1.
Support for the theory that meiosis arose from bacterial transformation comes from the increasing evidence that early diverging lineages of eukaryotes have the core genes for meiosis. This implies that the precursor to meiosis was already present in the bacterial ancestor of eukaryotes. For instance the common intestinal parasite Giardia intestinalis, a simple eukaryotic protozoan was, until recently, thought to be descended from an early diverging eukaryotic lineage that lacked sex. However, it has since been shown that G. intestinalis contains within its genome a core set of genes that function in meiosis, including five genes that function only in meiosis. In addition, G. intestinalis was recently found to undergo a specialized, sex-like process involving meiosis gene homologs. This evidence, and other similar examples, suggest that a primitive form of meiosis, was present in the common ancestor of all eukaryotes, an ancestor that arose from antecedent bacteria.
Origin and function Of Meiosis and Mitosis
Meiosis is ubiquitous among eukaryotes. It occurs in single-celled organisms such as yeast, as well as in multicellular organisms, such as humans. Eukaryotes arose from prokaryotes more than 1.5 billion years ago, and the earliest eukaryotes were likely single-celled organisms. To understand meiosis in eukaryotes, it is necessary to understand
(1) How meiosis arose in single celled eukaryotes,
(2) The function of Meiosis and Mitosis
1) Origin
There are two conflicting theories on how meiosis arose. One is that meiosis evolved from bacterial sex (called transformation) during the evolution of eukaryotes. The other is that meiosis arose from mitosis.
(1) How meiosis arose in single celled eukaryotes,
(2) The function of Meiosis and Mitosis
1) Origin
There are two conflicting theories on how meiosis arose. One is that meiosis evolved from bacterial sex (called transformation) during the evolution of eukaryotes. The other is that meiosis arose from mitosis.
- Theory that meiosis evolved from bacterial sex(transformation)
- Theory that meiosis evolved from mitosis
- Sharing of components during the evolution of meiosis and mitosis
2) Function
Single-celled eukaryotes (protists) generally can reproduce asexually (vegetative reproduction) or sexually, depending on conditions. Asexual reproduction involves mitosis, and sexual reproduction involves meiosis. When sex is not an obligate part of reproduction, it is referred to as facultative sex. Present-day protists, generally, are facultative sexual organisms, as are many bacteria. The earliest form of sexual reproduction in eukaryotes was probably facultative, like that of present-day protists. To understand the function of meiosis in facultative sexual protists, we next consider under what circumstances these organisms switch from asexual to sexual reproduction, and what function this transition may serve.Genetic Engineering Structures
Genetic engineering : Definition
Definition:
Definition:
Genetic engineering, also called genetic modification is the direct manipulation of the genome of an organism using biotechnology. (Genetic Modification indirectly through artificial selection has been practiced for centuries.) New DNA can be inserted into the host genome by first isolating and copy the genetic material of interest using molecular cloning methods to generate a DNA sequence, or by DNA synthesis, and then inserting this construct into the host organism. Genes can be deleted or "knocked out" using a nuclease. Gene targeting is a different technique that uses homologous recombination to change an endogenous gene, and can be used to delete a gene, exons remove, add a gene, or to introduce mutations.
An organism that is generated through genetic engineering is considered to be a genetically modified organism (GMO). The first GMOs were bacteria in 1973, transgenic mice were generated in 1974. Insulin-producing bacteria were commercialized in 1982 and genetically modified food has been sold since 1994. Glofish, the first GMO designed as a pet, was first sold in the United States in December 2003. [1]
Genetic engineering techniques have been applied in many fields, such as research, agriculture, industrial biotechnology, and medicine. Enzymes are used in detergents and medicines such as insulin and human growth hormone are produced in genetically modified cells, cell lines genetically modified experimental and transgenic animals such as mice or zebrafish are used for research purposes, and GM crops have been commercialized.
Structures Of Genetic Engineering: DNA Molecule
Definition of Genetic engineering & Genetic Engineering Structures
Definition
Genetic engineering alters the genetic makeup of an organism using techniques that remove heritable material or that introduce DNA prepared outside the organism either directly into the host or into a cell that is then fused or hybridized with the host. This involves using recombination nucleic acid (DNA or RNA) techniques to form new combinations of heritable genetic material followed by the incorporation of that material either indirectly through a vector system or directly through micro-injection, macro-injection and micro-encapsulation techniques. Genetic engineering does not normally include traditional animal and plant breeding, in vitrofertilisation, induction of polyploidy, mutagenesis and cell fusion techniques that do not use recombination nucleic acids or a genetically modified organism in the process. However the European Commission has also defined genetic engineering broadly as including selective breeding and other means of artificial selection. Cloning and stem cell research, although not considered genetic engineering, are closely related and genetic engineering can be used within them. Synthetic biology is an emerging discipline that takes genetic engineering a step further by introducing artificially synthesized genetic material from raw materials into an organism.
If genetic material from another species is added to the host, the resulting organism is called transgenic. If genetic material from the same species or a species that can naturally breed with the host is used the resulting organism is called cisgenic. Genetic engineering can also be used to remove genetic material from the target organism, creating a gene knockout organism.[8] In Europe genetic modification is synonymous with genetic engineering while within the United States of America it can also refer to conventional breeding methods.[9][10] The Canadian regulatory system is based on whether a product has novel features regardless of method of origin. In other words, a product is regulated as genetically modified if it carries some trait not previously found in the species whether it was generated using traditional breeding methods (e.g., selective breeding, cell fusion, mutation breeding) or genetic engineering.[Within the scientific community, the term genetic engineering is not commonly used; more specific terms such as transgenic are preferred.
Structures Of Genetic Engineering
Genetic engineering : Definition
Definition:
Definition:
Genetic engineering, also called genetic modification is the direct manipulation of the genome of an organism using biotechnology. (Genetic Modification indirectly through artificial selection has been practiced for centuries.) New DNA can be inserted into the host genome by first isolating and copy the genetic material of interest using molecular cloning methods to generate a DNA sequence, or by DNA synthesis, and then inserting this construct into the host organism. Genes can be deleted or "knocked out" using a nuclease. Gene targeting is a different technique that uses homologous recombination to change an endogenous gene, and can be used to delete a gene, exons remove, add a gene, or to introduce mutations.
An organism that is generated through genetic engineering is considered to be a genetically modified organism (GMO). The first GMOs were bacteria in 1973, transgenic mice were generated in 1974. Insulin-producing bacteria were commercialized in 1982 and genetically modified food has been sold since 1994. Glofish, the first GMO designed as a pet, was first sold in the United States in December 2003. [1]
Genetic engineering techniques have been applied in many fields, such as research, agriculture, industrial biotechnology, and medicine. Enzymes are used in detergents and medicines such as insulin and human growth hormone are produced in genetically modified cells, cell lines genetically modified experimental and transgenic animals such as mice or zebrafish are used for research purposes, and GM crops have been commercialized.
Cutting And Joining Of DNA Molecules
Cutting and Joining DNA Molecules
CUTTING DNA MOLECULES :
CUTTING DNA MOLECULES
It is worth recalling that prior to 1970 there was simply no method available for cutting a duplex DNA \molecule into discrete fragments. DNA biochemistry was circumscribed by this impasse. It became apparent that the related phenomena of host-controlled restriction and modification might lead towards a solution to the problem when it was discovered that restriction involves specific endonucleases. The favourite organism of molecular biologists, E. coil K12, was the first to be studied in this regard, but turned out to be an unfortunate choice. Its endonuclease is perverse in the complexity of its behaviour. The breakthrough in 1970 came with the discovery in Hae,nophilus influenzae of an enzyme that behaves more simply. Present-day DNA technology is totally dependent upon our ability to cut DNA molecules at specific sites with restriction endonucleases. An account of host-controlled restriction and modification therefore forms the first part of this chapter.
higher order chromatin organization 2013 | Higher order organization of eukaryotic chromatin
Higher order organization of eukaryotic chromatin :
In 1974, Roger Kornberg on the basis of micro coccal nuclease digestion pattern of genomic DNA proposed that the lowest level of chromatin organization consists of wrapping of about 200 bp of DNA double helix around a histone octamer. In strict sense, about 147 bp or 1 and ¾ turn of DNA is wound around a block of two copies of histone proteins. H2A, H2B, H3 and H4 each and constitutes the nucleosome core particle. These nucleosomes are connected to each other like beads of pearls on a string through variable length of linker DNA. Recent observations on x-ray crystallographic structure of nucleosome reveal that the architectural motif of nucleosome consists of I,islone folds having (H3),-(l-14), tetramer at the centre and H2A and H2B dimers at the end of the length. The core fold contains 121 bp DNA. Rest 26 bp DNA is contained by N-terminal end of histone units, having about 13 bp at each end. Histone proteins interact with phosphodiestcr backbone and deoxy ribose moiety at minor grooves of DNA
through electrostatic interactions and hydrogen bonding, but do not have any base specific contacts. Each of the histone unit has 10-40 amino acid long N terminal tail sections which can be extended, remains outside the core section and plays important role in chromatin modeling and transcriptional regulation. Another histone protein HI and H5 (in birds) serves as the linker histone by binding to the nucleosome near one end of core DNA and influence the entry and exit of DNA into nucleosome. HI was considered to stabilize additional 10 bp DNA at each end, thereby putting 167 (147 +10+10) bp DNA around nucleosome. The
assembly of 167 bp DNA, Hi and core histone octamer is sometimes referred as chronzatosome. However, recent studies reveal that the repeating unit of higher order chromatin structure contains 156-157 bp DNA in the nucleosome and is known as the 10 nm fiber of DNA due to the nucleosome unit diameter of 10 nm. Nucleosomes are often positioned in specific genomic regions (promoters, regulatory elements, specific sequences etc.) which is gene sequence dependant, a phenomenon known as nucleosome positioning that is suggested to be involved in regulation of gene expression. It has been observed that this positioning is not static; rather nucleosomes remain in a dynamic state in the chromatin.This preferential positioning helps nucleosome to repress transcription. The flexible tails are basic in nature and serve as substrates for many enzymes involved in methylation, acetylation, phosphorylation and other post-translational modifications. This
has led to the birth of a his tone code hypothesis which postulates that the specific arrangements and sequences of histone tail are read as a code to unfold and remodel chrornatin, and in gene expression. It has also been observed that the core histone proteins are also extensively modified and thereby can also be viewed as apart of histone code. Two types of posttranslational modifications, class I and class II modifications are suggested. Class I is involved in histonc-non histone chromatin associated protein interaction, while class II involves histone-DNA and histone-histone interactions.
Second level of chromatin organization constitutes of 30 nm fiber, as observed as a filament of diameter 30 nm under electron microscope. It is a package of t of nucleosomes and is maintained by inter histone interactions, which can be classified into eight distinct types. The interaction between N terminal of histone H4 and part of H2A-H2B dimer is essential for maintenance of 30 nm structure. Previous idea that the 30 nm fiber comprises of an one start solenoid structure of 6-8 nucleosome units is being challenged by recent microscopic and crystallographic observations. Recent findings suggested that 30 nm fiber may be comprised of stacks of nucleosomes arranged in a hc’o start helical model or one start helical imiodel with a very high nucleosome packing ratio (15-22 nucleosomes per unit instead of 6-8 in solenoid model).

Still, the packing ratio, 6 for each nucleosome and about 120 for each 30 nm fiber unit having 20 nucleosomes each (20 x 6) is insufficient for packing DNA into small nucleus. The 30 nm fiber again folds and refolds to form loops of 40-90 kb and the torsion created therein is relieved by action of topoisomerases. The higher order nuclear scaffold structures are considered to be held by structural maintenance proteins (SMCs). Theses proteins also play major role in holding of two chromatids during divisional phases. Recent observations suggest that the organization of chromatin material in higher as well as lower eukaryotes is dynamic in nature in the sense that the individual chromosomes can reposition themselves in the nuclear interior. The chromosomes occupy specific regions in the nucleus known as chromosome territories with euchromatic regions oriented to the interior and heterochrornatic regions at the peripheral space. These chromatins can move within the nucleus to a certain extent, observed to be more at interphase. This spatial distribution and movement of chromatin material is suggested to provide a mechanism for controlling gene expression by allowing binding of genes with localized transcriptional factors, intcrchromosomal interactions between loci present on different chromosomes (gene kissing) and by inducing epigenetic regulations. A number of protein and RNA molecules are involved in prevention of movement of chrorna tin material (chroina tin immobilization), thereby imposing restriction on random movement of the chromosomes in the nucleus.Genome organization in eukaryotic chromosomes at metaphase as well as at interphase is non uniform. A chromosome based on metaphase morphology shows vanous landmarks with distinctive features. Tlie cent romere or primary constriction, where microtubules are attached during anaphasic movement constitutes mostly of hetcrochromatin and are attached to protein complexes known as kinetochore that act as a bridge between microtubules and centromere. For correct chromosome segregation it is e.ential that only one centromere he present in a chromosome. Size of a centromere in eukaryotes may vary greatly, from as little as 125 bp in yeast to several Mb in human. The two ends of the linear chromosomes are known as telo,neres, having complex looped structures involving non-Watson Crick base pairing, hcterochromatinized regions and unusual replication mechanism. A number of proteins arc associated with telomere to prevent DNA damage at chromosome ends. Between centromere and telomere, further constricted regions or secondary constrictions max’ exist. The area between a telomere and nearest secondary constrictions is sometimes called as satellite. Nucleolus is preferentially attached to a region of chromosome known as nuclolare organiser region (NOR).
Diakinesis
Chromosomes condense further during the diakinesis stage, from Greek words meaning "moving through".30 This is the first point in meiosis where the four parts of the tetrads are actually visible. Sites of crossing over entangle together, effectively overlapping, making chiasmata clearly visible. Other than this observation, the rest of the stage closely resembles prometaphase of mitosis; the nucleoli disappear, the nuclear membrane disintegrates into vesicles, and the meiotic spindle begins to form.[citation needed]
Diplotene
During the diplotene stage, also known as diplonema, from Greek words meaning "two threads",30 the synaptonemal complex degrades and homologous chromosomes separate from one another a little. The chromosomes themselves uncoil a bit, allowing some transcription of DNA. However, the homologous chromosomes of each bivalent remain tightly bound at chiasmata, the regions where crossing-over occurred. The chiasmata remain on the chromosomes until they are severed in anaphase I.[citation needed]
In human fetal oogenesis all developing oocytes develop to this stage and stop before birth. This suspended state is referred to as the dictyotene stage and remains so until puberty.
Pachytene
The pachytene stage, also known as pachynema, from Greek words meaning "thick threads",:27 is the stage when chromosomal crossover (crossing over) occurs. Nonsister chromatids of homologous chromosomes may exchange segments over regions of homology. Sex chromosomes, however, are not wholly identical, and only exchange information over a small region of homology. At the sites where exchange happens, chiasmata form. The exchange of information between the non-sister chromatids results in a recombination of information; each chromosome has the complete set of information it had before, and there are no gaps formed as a result of the process. Because the chromosomes cannot be distinguished in the synaptonemal complex, the actual act of crossing over is not perceivable through the microscope, and chiasmata are not visible until the next stage.[citation needed]
Zygotene
The zygotene stage, also known as zygonema, from Greek words meaning "paired threads",27 occurs as the chromosomes approximately line up with each other into homologous chromosome pairs. This is called the bouquet stage because of the way the telomeres cluster at one end of the nucleus. At this stage, the synapsis (pairing/coming together) of homologous chromosomes takes place, facilitated by assembly of central element of the synaptonemal complex. Pairing is brought about in a zipper-like fashion and may start at the centromere (procentric), at the chromosome ends (proterminal), or at any other portion (intermediate). Individuals of a pair are equal in length and in position of the centromere. Thus pairing is highly specific and exact. The paired chromosomes are called bivalent or tetrad chromosomes.[citation needed]
Leptotene
The first stage of prophase I is the leptotene stage, also known as leptonema, from Greek words meaning "thin threads".[8]:27In this stage of prophase I, individual chromosomes—each consisting of two sister chromatids—change from the diffuse state they exist in during the cell's period of growth and gene expression, and condense into visible strands within the nucleus.27:353 However the two sister chromatids are still so tightly bound that they are indistinguishable from one another. During leptotene, lateral elements of the synaptonemal complex assemble. Leptotene is of very short duration and progressive condensation and coiling of chromosome fibers takes place.
Prophase I
It is the longest phase of meiosis. During prophase I, DNA is exchanged between homologous chromosomes in a process called homologous recombination. This often results in chromosomal crossover. The new combinations of DNA created during crossover are a significant source of genetic variation, and may result in beneficial new combinations of alleles. The paired and replicated chromosomes are called bivalents or tetrads, which have two chromosomes and four chromatids, with one chromosome coming from each parent. The process of pairing the homologous chromosomes is called synapsis. At this stage, non-sister chromatids may cross-over at points called chiasmata (plural; singular chiasma).
Meiosis I
Meiosis I separates homologous chromosomes, producing two haploid cells (N chromosomes, 23 in humans), and thus meiosis I is referred to as a reductional division. A regular diploid human cell contains 46 chromosomes and is considered 2N because it contains 23 pairs of homologous chromosomes. However, after meiosis I, although the cell contains 46 chromatids, it is only considered as being N, with 23 chromosomes. This is because later, in Anaphase I, the sister chromatids will remain together as the spindle fibers pull the pair toward the pole of the new cell. In meiosis II, an equational division similar to mitosis will occur whereby the sister chromatids are finally split, creating a total of 4 haploid cells (23 chromosomes, N) - two from each daughter cell from the first division.
Phases Of Meiosis
Meiosis is divided into meiosis I and meiosis II which are further divided into Karyokinesis I and Cytokinesis I & Karyokinesis II and Cytokinesis II respectively.
1st Phases Of Meiosis
Synchronous processes
2nd Phases Of Meiosis
1st Phases Of Meiosis
Synchronous processes
2nd Phases Of Meiosis
- Meiosis II
Subscribe to:
Comments (Atom)